MichaelisMenten plot for enzymesubstrate Download Scientific Diagram
The Michaelis-Menten mechanism (Michaelis & Menten, 1913) is one which many enzyme mitigated reactions follow. The basic mechanism involves an enzyme ( E, a biological catalyst) and a substrate ( S) which must connect to form an enzyme-substrate complex ( ES) in order for the substrate to be degraded (or augmented) to form a product ( P ).
MichaelisMentenKonstante · Einfach erklärt Enzymaktivität [mit Video] · [mit Video]
In biochemistry, the direct linear plot is a graphical method for enzyme kinetics data following the Michaelis-Menten equation. [1] In this plot, observations are not plotted as points, but as lines in parameter space with axes and , such that each observation of a rate at substrate concentration is represented by a straight line with.

MichaelisMenten study of the nonenzymatic RNA copying rates of 2AIpG... Download Scientific
The Michaelis-Menten mechanism (Michaelis & Menten, 1913) is one which many enzyme mitigated reactions follow. The basic mechanism involves an enzyme ( E E, a biological catalyst) and a substrate ( S S) which must connect to form an enzyme-substrate complex ( ES E S) in order for the substrate to be degraded (or augmented) to form a product ( P.

MichaelisMenten MichaelisMenten plot of MpIspS initial... Download Scientific Diagram
The Michaelis-Menten equation (see below) is commonly used to study the kinetics of reaction catalysis by enzymes as well as the kinetics of transport by transporters. Typically, the rate of reaction (or reaction velocity) is experimentally measured at several substrate concentration values. The range of substrate concentrations is chosen such that very low reaction rates as well as saturating.

Image result for michaelis menten graph Reaction Rate, Biochemical, Biochemistry, Line Chart
Michaelis Menten function to describe relation of concentration of substrate and enzyme velocity. Sample Curve Parameters. Number: 2 Names: Vmax, Km Meanings: Vmax = Maximum Velocity, Km = Michaelis Constant Lower Bounds: Vmax > 0.0, Km > 0.0 Upper Bounds: none Script Access nlf_MichaelisMenten (x,Vmax,Km) Function File. FITFUNC\MichaelisMenten.FDF

Example of a MichaelisMenten plot (left) and a LineweaverBurk plot Download Scientific Diagram
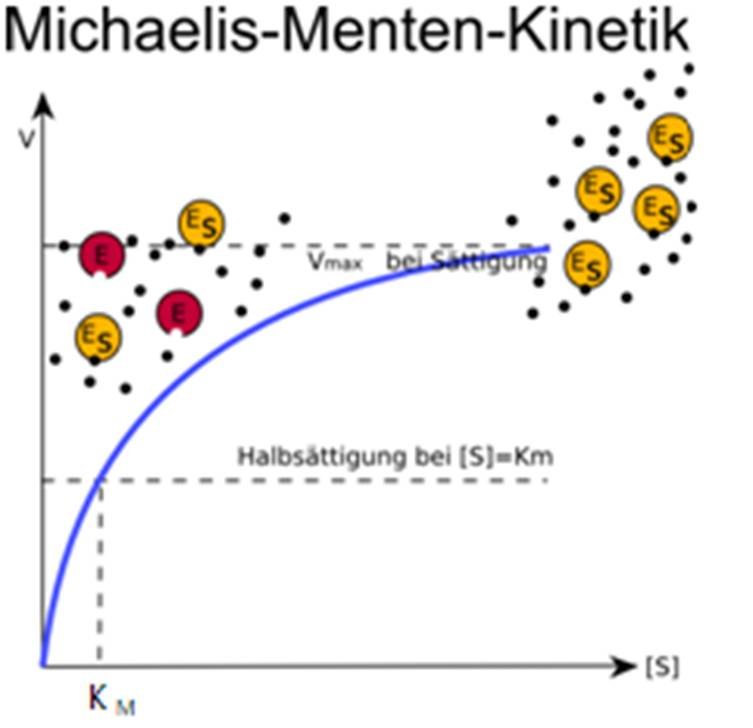
Page ID. Two 20 th century scientists, Leonor Michaelis and Maud Leonora Menten, proposed the model known as Michaelis-Menten Kinetics to account for enzymatic dynamics. The model serves to explain how an enzyme can cause kinetic rate enhancement of a reaction and explains how reaction rates depends on the concentration of enzyme and substrate.

MichaelisMenten of αglucosidase activity, and in the... Download Scientific Diagram
Michaelis-Menten derivation for simple steady-state kinetics. The Michaelis-Menten equation is a mathematical model that is used to analyze simple kinetic data.The model has certain assumptions, and as long as these assumptions are correct, it will accurately model your experimental data.The derivation of the model will highlight these assumptions.

MichaelisMenten enzyme for TMT1 (green diamonds ), TMT2... Download Scientific Diagram
Michaelis and Menten equation (MM equation) has dominated biochemistry for more than a century after its seminal introduction in a paper published in 1913 in the journal Biochemische Zeitschrift, a predecessor of FEBS Journal [[]].Hence, publishing this guide in FEBS Journal would represent an apt dedication to the unmatched service rendered by this journal to the scientific community for more.

The Michaelis Menten plot for HRP to determine K m and V max of the... Download Scientific
The Michaelis constant is the substrate concentration at which an enzyme operates at one half of its maximum velocity. How can you calculate this constant ba.

The MichaelisMenten Enzyme Model
This short video shows you how to set up the Michaelis-Menten model in Excel to model the creation of product as a function of substrate concentration.
MichaelisMenten YouTube
The program determines the constants Rmax and Km of the Michaelis-Menten model using data given in Fundamentals of Chemical Reaction Engineering by M. E. Davis & R. J. Davis, McGraw Hill, 2003. This data represents the substrate (catechol) concentration versus time. This data was obtained using E. herbicola immobilized in a polymer gel to.

The MichaelisMenten diagram of soluble and immobilized form of DHODH... Download Scientific
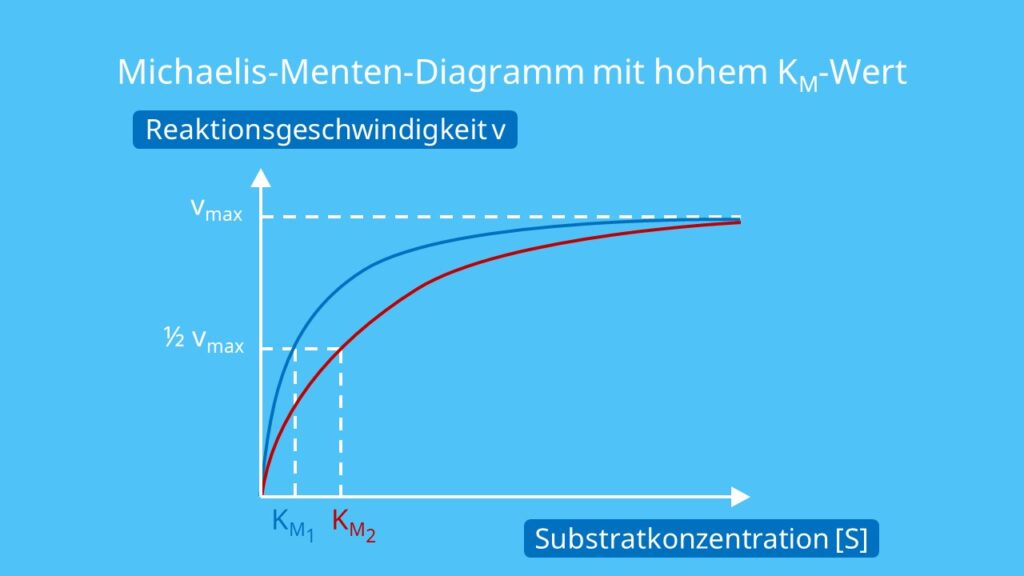
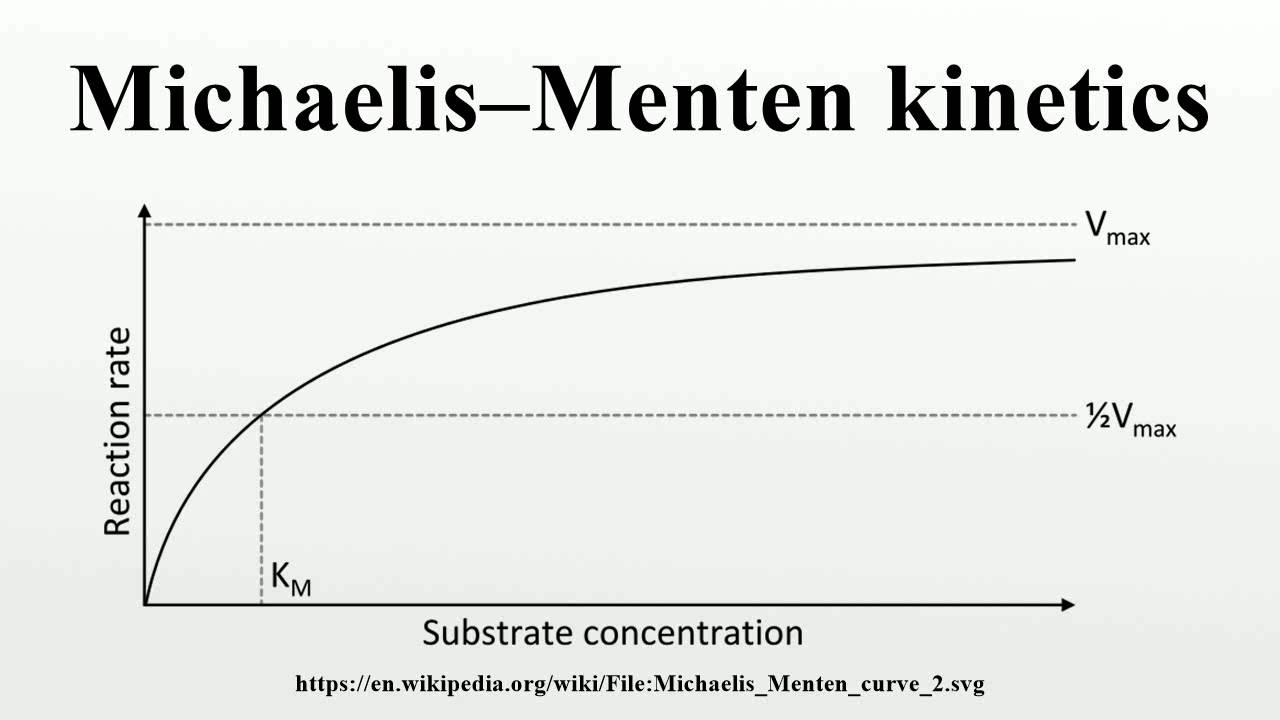
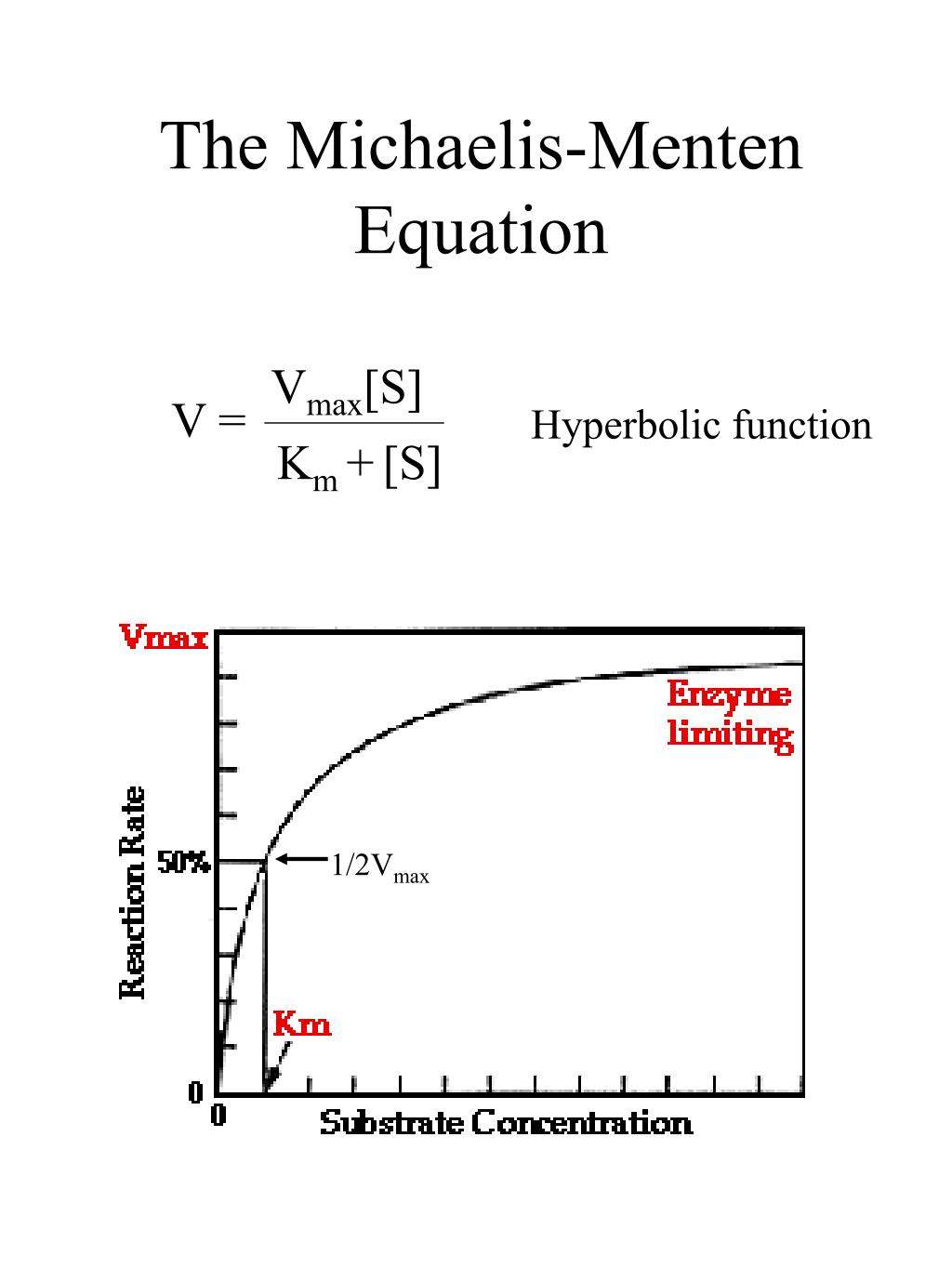
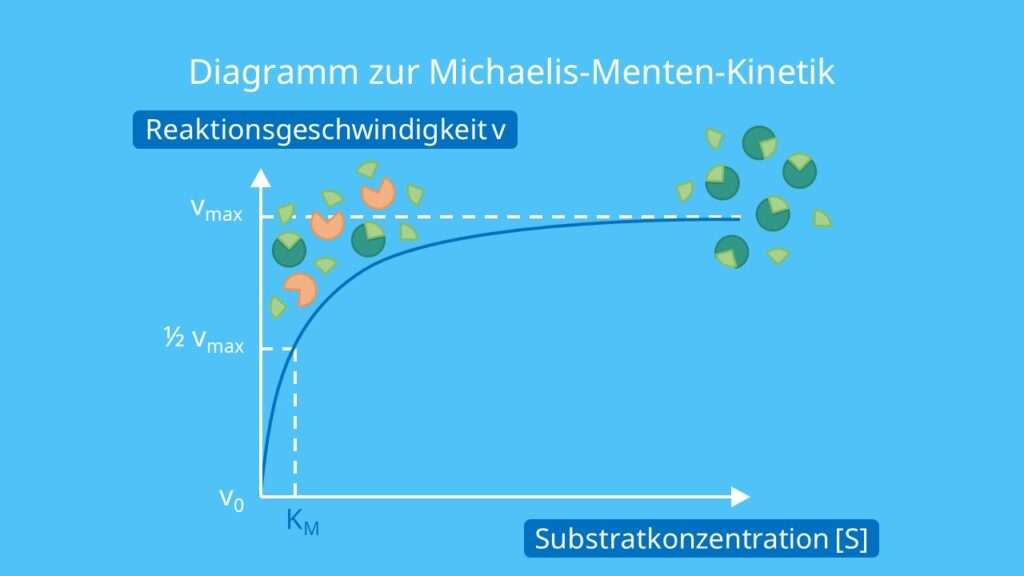
The reaction is zero-order kinetics. Figure 5.3.2: Diagram of reaction velocity and Michaelis-Menten kinetics. v = Vmax 2 = Vmax[S] Km + [S] (5.3.17) (5.3.17) v = V m a x 2 = V m a x [ S] K m + [ S] Therefore, Km K m is equal to the concentration of the substrate when the rate is half of the maximum velocity.

MichaelisMentenGleichung… BASICS of ANESTHESIOLOGY boa.coach
Step 4: Format the Scatter Plot. Next, format the scatter plot to turn it into a Michaelis-Menten plot. First, remove the vertical axis and format the horizontal axis to become a logarithmic scale. This can be done by right-clicking the horizontal axis and selecting "Format Axis.". Then, select "Logarithmic Scale" under the "Scale.
Physikalische Chemie
Michaelis-Menten kinetics. In biochemistry, Michaelis-Menten kinetics, named after Leonor Michaelis and Maud Menten, is the simplest case of enzyme kinetics, applied to enzyme-catalysed reactions of one substrate and one product. It takes the form of an equation describing the reaction rate (rate of formation of product P, with.
PPT LAB 3 Enzyme PowerPoint Presentation, free download ID4526880
The Michaelis-Menten equation, named after biochemist Leonor Michaelis and physician Maud Menten, "describes the relationship between the rate of substrate conversion by an enzyme (V) and the concentration of the substrate ([S])," according to the Davidson College Chemistry website. Based on this equation, the Michaelis-Menten curve can be.
MichaelisMentenGleichung · einfach erklärt Formel [mit Video] · [mit Video]
Km is the Michaelis-Menten constant, in the same units as X. It is the substrate concentration needed to achieve a half-maximum enzyme velocity. Create a Lineweaver-Burk plot. Before nonlinear regression was available, investigators had to transform curved data into straight lines, so they could analyze with linear regression. One way to do.